-
Notifications
You must be signed in to change notification settings - Fork 18
MRI_Opera_export_tools
The tool stitches images from the Opera Phenix HCS System. It reads the Index.idx.xml file to pre-arrange the images and then stitches and fuses them using the Grid/Collection stitching-plugin [1]. Images are stitched by plane and channel. Z-stacks and multi-channel images can optionally be created. Projections can also be created.
The source code in git-hub can be found here.
To install the tool save the files opera_export_tools.ijm and opera_export_tools.py into the folder macros/toolsets of your FIJI installation.
Select the "opera_export_tools" toolset from the >> button of the ImageJ launcher.
- the first button opens this help page
- second button allows to select the index-file
- the third button allows to select the wells, that will be exported
- the fourth button allows to modify the export options
- the fifth button starts the batch-export
Press the Set index-file-button and navigate to the folder containing the images. Select the index file Index.idx.xml.
The path must not contain spaces or special characters such as accents. If necessary rename folders before selecting the index-file!
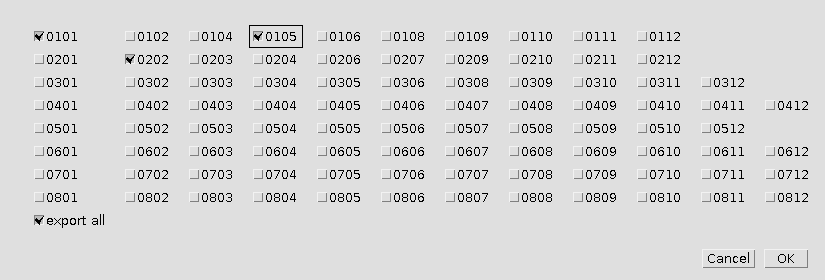
Press the Select wells-button and select the wells you want to export. If export all is selected, all wells will be exported.
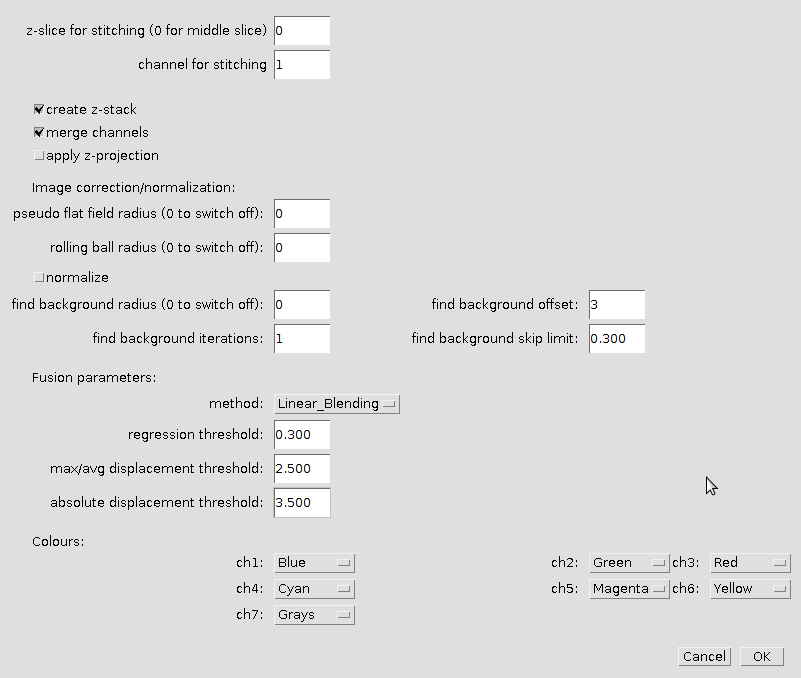
Press Set options-button.
- z-slice for stitching
- The z-slice, that is used to calculate the stitching. Enter 0 to use the middle slice.
- channel for stitching
- The channel, that is used to calculate the stitching
- create z-stack
- If selected one file per z-stack is created, otherwise one per slice is created.
- merge channels
- If selected the channels are merged into a hyperstack-image.
- apply z-projection
- If selected a maximum intensity projection (mip) is created instead of a z-stack.
- pseudo flat field radius
- The radius for the pseudo flat field correction. Set the radius to 0 if you do not want to apply a pseudo flat field correction. You must have the biovoxxel toolbox [2] installed
- rolling ball radius
- The radius for the rolling-ball background correction. Set the radius to 0 if you do not want to apply a rolling-ball background correction.
- normalize
- If selected the the intensities of the tiles of a mosaic are normalized.
- find background radius
- The radius for the find and subtract background operation. Set the radius to 0 if you do not want to apply the find and subtract background operation.
- Fusion parameters:
- The parameters for the stitching plugin [1]
- Colours
- Set the colours of the first seven channels.
Press the Launch export-button to start the batch export of the images.
The result images will be written into a subfolder out of the folder that contains the index-file.
-
S. Preibisch, S. Saalfeld, P. Tomancak (2009) “Globally optimal stitching of tiled 3D microscopic image acquisitions”, Bioinformatics, 25(11):1463-1465. Webpage PDF
-
The BioVoxxel Image Processing and Analysis Toolbox. Brocher, 2015, EuBIAS-Conference, 2015, Jan 5

 Volker Bäcker
Volker Bäcker